

Discovery Boulevard
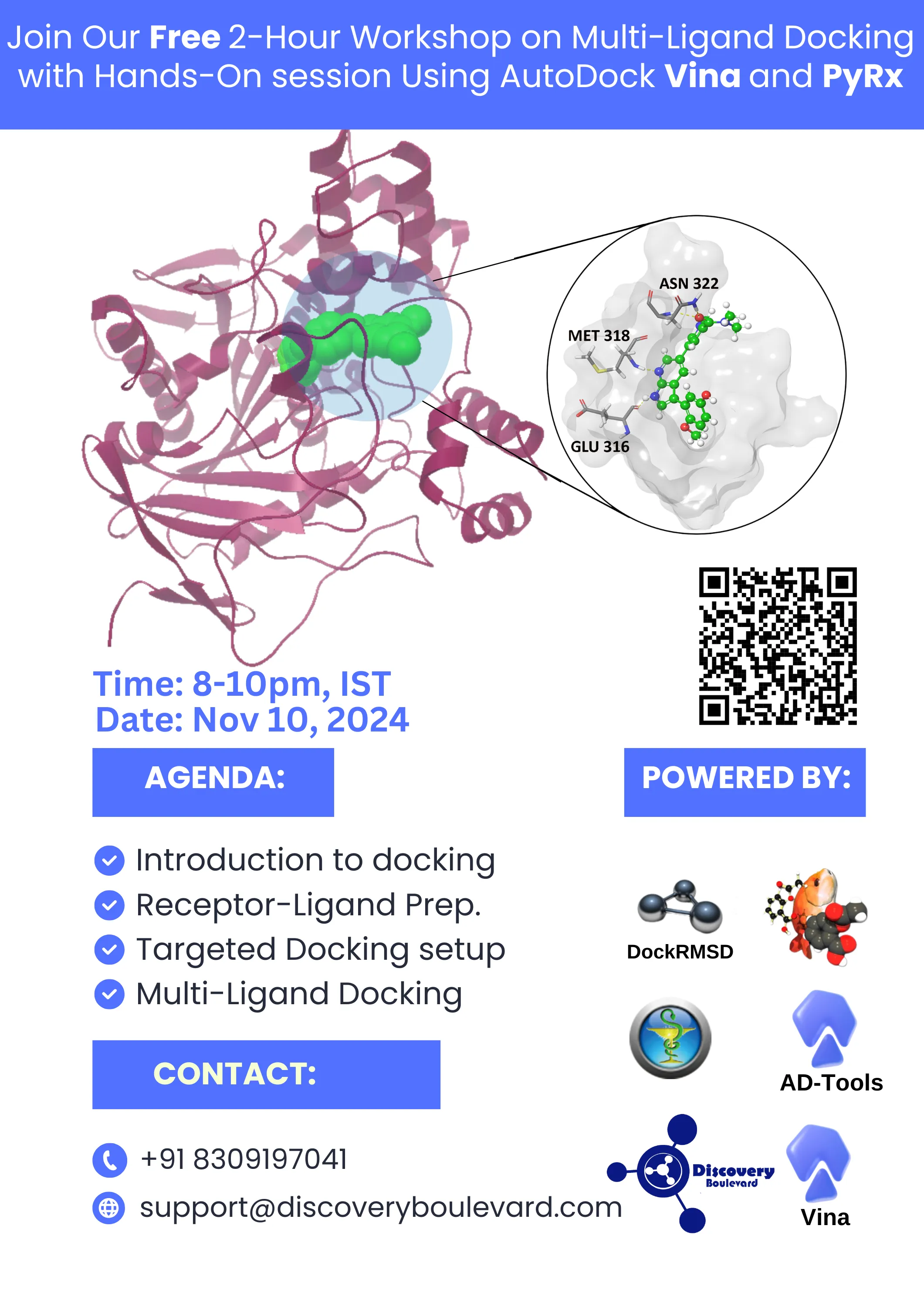
2-Hour Hands-On Multi-Ligand Docking with AutoDock Vina and PyRx
Hosted by Discovery Boulevard
Discovery Boulevard

NOV
10
Sun, 10 Nov
02:30 PM - 04:30 PM (Coordinated Universal Time)
Online
Register to get link
Event starts in
0
days
0
hours
0
min
0
sec
Get your tickets



Ticket Price$5
We’re full! Check back soon.
Ticket Price$5
We’re full! Check back soon.
About
Agenda:
1. Introduction to Multi-Ligand Docking
Overview of docking multiple ligands against a single receptor.
Introduction to AutoDock Vina and its advantages in high-throughput docking.
2. Selecting and Preparing the Target Receptor:
Downloading the receptor structure from the PDB database.
Cleaning and preparing the receptor (removing water molecules, adding hydrogens, adding charge, etc.).
3. Converting to. pdbqt format:
Using AutoDockTools (ADT) to convert the receptor to. pdbqt format.
Setting up the active site by defining the grid box around the binding site.
4. Creating or Downloading a Virtual Ligand Library:
Downloading ligands from the ZINC database.
Ensuring ligands are energy minimized and in the correct format.
Preparing multiple ligands for docking (converting to .pdbqt using Open Babel).
5. Configuration of Vina Input Files:
Preparing a configuration file (.conf) for AutoDock Vina specifying the receptor, grid box dimensions, and number of ligands to dock.
6. Running the Docking:
Executing AutoDock Vina in command line using single ligand.
Multiple ligands docking using PyRx software.
7. Docking Results Overview:
Reviewing docking scores (binding affinities) for each ligand.
Extracting and analyzing the best binding modes and docking scores from the output files.
Sorting ligands based on their binding affinity and energy scores.
Selecting the top candidates for further analysis.
8. Visualizing Binding Modes:
Using Maestro or Online tools to visualize the ligand-receptor interactions.
Identifying key interactions (hydrogen bonds, hydrophobic interactions, pi-pi stacking, etc.).
9. Redocking and RMSD Validation:
Performing re-docking with the native ligand to validate the docking protocol.
Superimposing the docked poses with the native ligand to check RMSD values.
Event By
Ask a question
14 people attending





Location

Register to get event link
Online
This event is part of a community

Discovery Boulevard
By purchasing this, you also unlock
Community access
New updates
29 Posts
Exclusive content
Whatsapp Group
2-Hour Hands-On Multi-Ligand Docking with AutoDock Vina and PyRx
Sun, 10 November • 02:30PM